Manual 10 for MD++
Visualize Disregistry Vector
Keonwook Kang and Wei Cai
If you use ddrij to visualize or analyze disregistry vectors in your simulation results, you are asked to cite the following paper:
Keonwook Kang and Wei Cai, Int. J. of Plasticity 26 (2010) 1387
Compile execution file
$ make ddrij build=R SYS=gpp
Run example file
Identify bonds
$ bin/ddrij_cpp ./scripts/Examples/example09-disreg.tcl 0
Read/plot bonds
$ bin/ddrij_cpp ./scripts/Examples/example09-disreg.tcl 1
Read bonds and calculate the disregistry vector
$ bin/ddrij_cpp ./scripts/Examples/example09-disreg.tcl 2
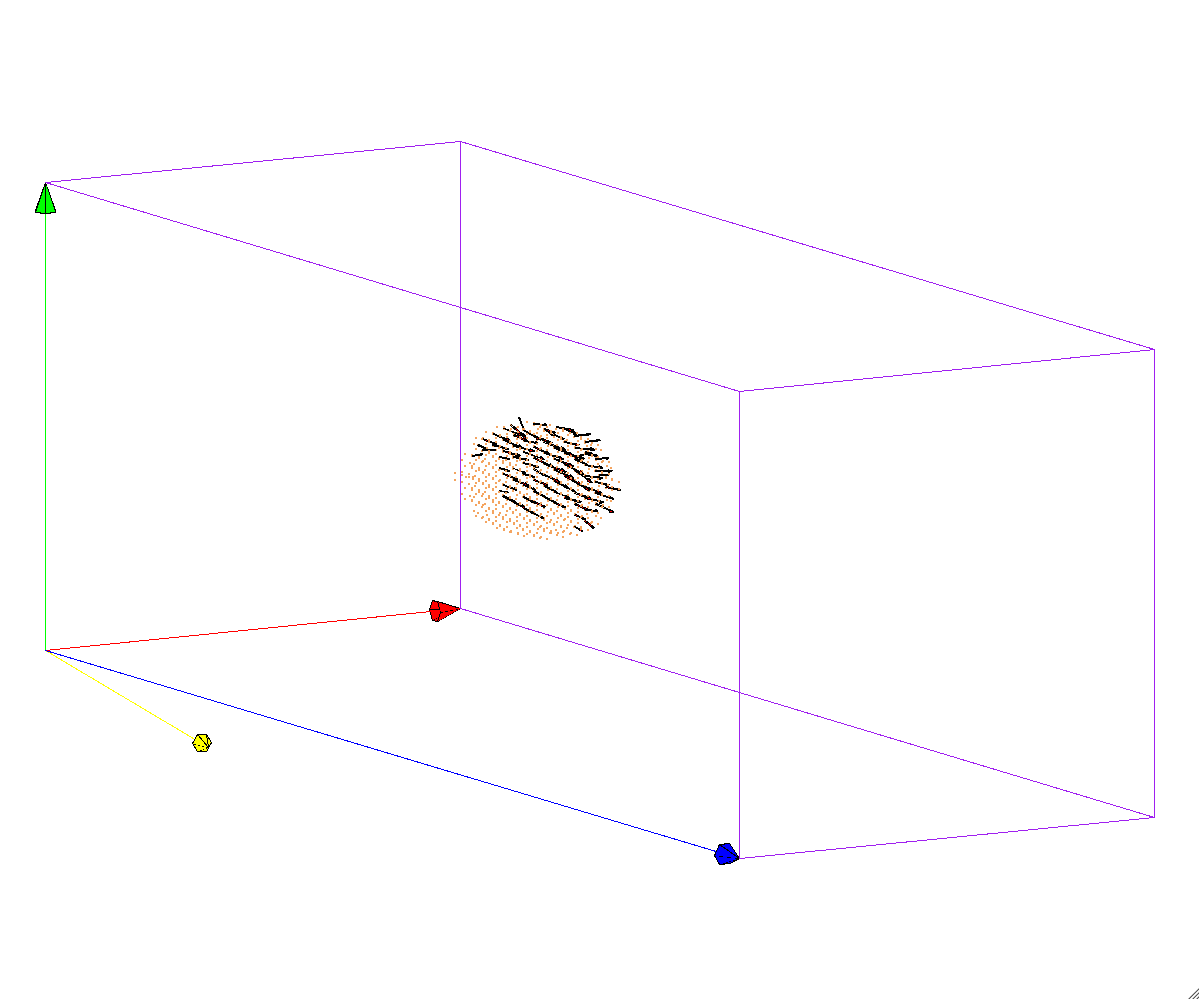
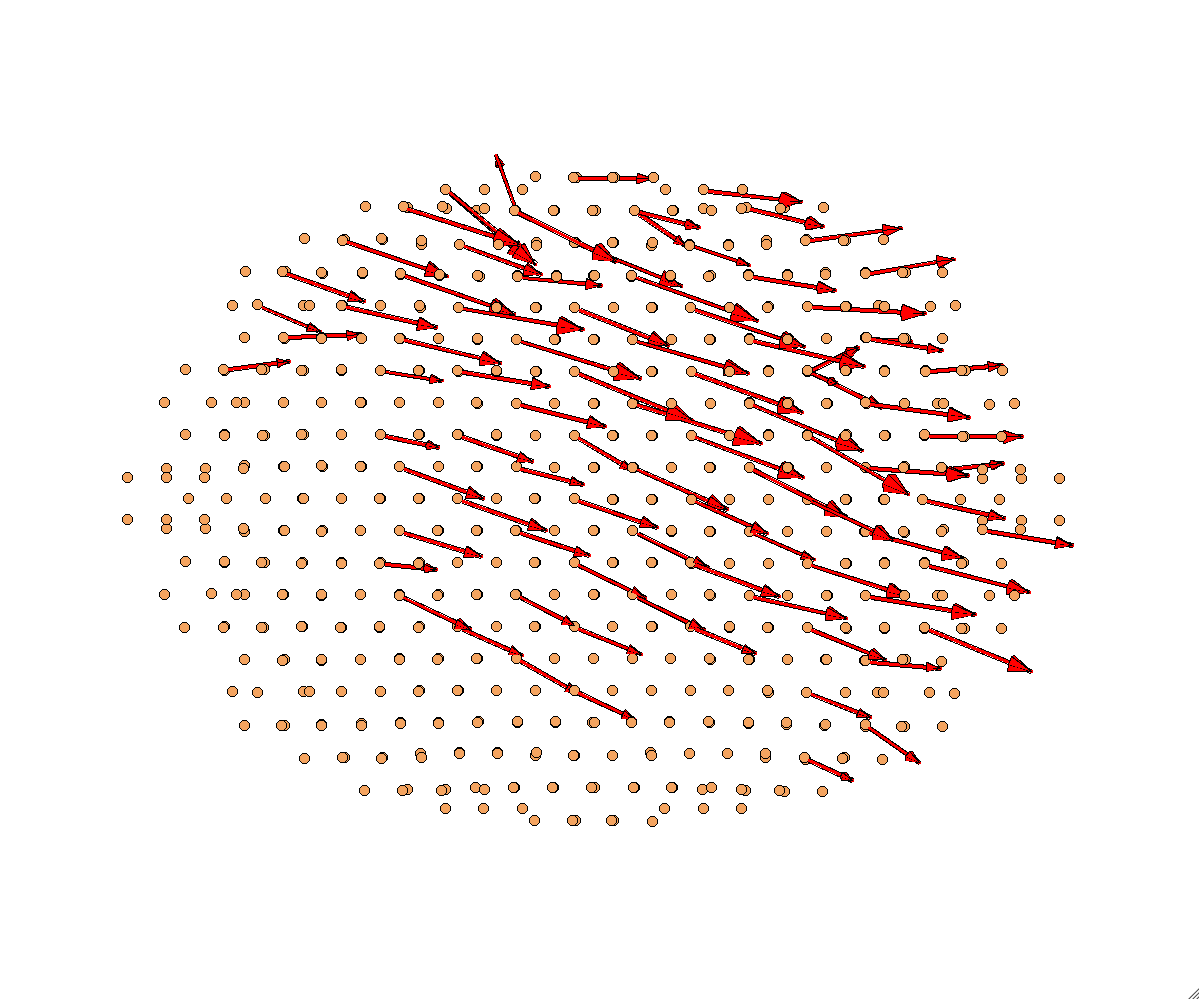
Plot disregistry vectors
$ bin/ddrij_cpp ./scripts/Examples/example09-disreg.tcl 3
# -*-shell-script-*-
source "scripts/Examples/Tcl/startup.tcl"
#*******************************************
# Definition of procedures
#*******************************************
proc initmd { n } {
MD++ setoverwrite
MD++ dirname = "runs/test-disreg"
MD++ NIC = 200 NNM = 400
}
proc DotProd {a b} {
# Calculate dot product of a and b
if {[llength $a] < 3 || [llength $b] < 3} {
return -code error "Vectors must have 3 components!"
}
scan $a "%f %f %f" ax ay az
scan $b "%f %f %f" bx by bz
return [format %22.16e [expr $ax*$bx + $ay*$by + $az*$bz]]
}
proc VectorNorm { a } {
# Calculate |a|.
if { [llength $a] < 3 } {
return -code error "Vectors must have 3 components!"
}
#scan $a "%f %f %f" ax ay az
set ax [lindex $a 0]; set ay [lindex $a 1]; set az [lindex $a 2];
set d [format %22.16e [expr sqrt($ax*$ax + $ay*$ay + $az*$az)]]
return $d
}
proc normalize {v} {
if {[llength $v] < 3 } {
return -code error "Vectors must have 3 components!"
}
set vx [lindex $v 0]; set vy [lindex $v 1]; set vz [lindex $v 2]
#set v2 [format %22.16e [expr $vx*$vx + $vy*$vy + $vz*$vz]]
#set v_mag [format %22.16e [expr sqrt($v2)]]
set v_mag [VectorNorm $v]
set ex [format %22.16e [expr $vx/$v_mag]]
set ey [format %22.16e [expr $vy/$v_mag]]
set ez [format %22.16e [expr $vz/$v_mag]]
return [list $ex $ey $ez]
}
proc fix_atoms_on_tilted_plane { sx0 sy0 sz0 d } {
#------------------------------------------------------------
#Select atoms to fix for dislocation nucleation
#
set C1 {0 0 1}; set C2 {1 -1 0}; set C3 {1 1 0}
#set n { 1 1 -1};
set n { 0.85 0.85 -1};
set e1 [normalize $C1]; set e2 [normalize $C2]; set e3 [normalize $C3]
set nx [DotProd $n $e1]; set ny [DotProd $n $e2]; set nz [DotProd $n $e3]
set n [list $nx $ny $nz];
puts "n = ([format %10.6e $nx], [format %10.6e $ny], [format %10.6e $nz])\
in unit of a in lab coordinate."
set NP [MD++_Get NP]
set Lx [MD++_Get H_11]; set Ly [MD++_Get H_22]; set Lz [MD++_Get H_33]
set x0 [expr $sx0*$Lx]; set y0 [expr $sy0*$Ly]; set z0 [expr $sz0*$Lz];
set N_fixed 0
for { set i 0 } { $i < $NP } { incr i 1 } {
set sx [MD++_Get SR( [expr $i*3 ] ) ]
set sy [MD++_Get SR( [expr $i*3+1] ) ]
set sz [MD++_Get SR( [expr $i*3+2] ) ]
set x [expr $Lx*$sx ]
set y [expr $Ly*$sy ]
set z [expr $Lz*$sz ]
#puts "atom ($i) ($x $y $z)"
set ndotx [expr ($x-$x0)*$nx + ($y-$y0)*$ny + ($z-$z0)*$nz]
if { [expr abs($ndotx)] <= $d } {
MD++ fixed($i) = 1
set N_fixed [expr $N_fixed + 1]
}
}
puts "Number of fixed atoms : $N_fixed"
}
proc setup_window { } { MD++ {
# Plot Configuration
atomradius = [0.3 0.4] bondradius = 0.3
atomcolor0 = SandyBrown atomcolor1 = LightGrey
bondcolor = red backgroundcolor = white #gray70
fixatomcolor = yellow
color00 = "orange" color01 = "purple" color02 = "green"
color03 = "magenta" color04 = "cyan" color05 = "purple"
color06 = "gray80" color07 = "white"
plot_map_pbc = 1
plot_color_axis = 0
win_width = 1200 win_height = 1000
plotfreq = 10 rotateangles = [ 30 -10 0 1.25 ]
} }
proc openwindow { } {
setup_window
MD++ openwin alloccolors rotate saverot eval plot
#MD++ calcentralsymmetry input = \[ 0 20 100 \] GnuPlotHistogram
}
#*******************************************
# Main program starts here
#*******************************************
if { $argc == 0 } {
set status 0
} elseif { $argc > 0 } {
set status [lindex $argv 0]
}
puts "status = $status"
if { $argc >= 0 && $argc <= 1 } {
set n 0; set m 0
} elseif { $argc > 1 && $argc <=2 } {
set n [lindex $argv 1]; set m 0
} elseif { $argc > 2 } {
set n [lindex $argv 1]; set m [lindex $argv 2]
}
puts "n = $n, m = $m"
# lattice constant of DC Si
set a_Si 5.431
if { $status == 0 } {
# Identify bonds
MD++ setnolog
initmd " "
# cut-off distance to declare a bond between two atoms
# usu. the distance to the 1st neighbor
set rc [expr $a_Si*sqrt(3)/4]
# Add buffer distance.
set rc [expr 1.3*$rc]
MD++ RLIST = 4.1490
# Read the reference configuration
MD++ incnfile = ../../structures/Examples/ref-testdisreg.cn readcn eval
# Will be used to determine the bond direction.
# If dot(slip_normal,r_{ij}) < 0, then r_{ij} := -r_{ij}.
MD++ slip_normal = \[ -1.0 0.0 1.414214 \]
# Specify the region where the diregistry analysis is applied
MD++ plot_limits = \[1 -1 1 -1 1 -0.150 -0.031 \]
MD++ input = \[ $rc \] identify_bonds
# Write bond data:
# [bond.i bond.j bond.center.x bond.center.y bond.center.z bond.type]
MD++ finalcnfile = bonds.dat write_bonds
openwindow
MD++ sleep quit
} elseif { $status == 1 } {
# Read/Plob bonds
MD++ setnolog
initmd " "
MD++ RLIST = 4.1490
MD++ incnfile = ../../structures/Examples/ref-testdisreg.cn readcn eval
# Read bond data
MD++ incnfile = bonds.dat read_bonds
setup_window
MD++ plot_limits = \[1 -1 1 -1 1 -0.150 -0.031 \]
MD++ openwin alloccolors rotate saverot eval plot_bonds
MD++ sleep quit
} elseif { $status == 2 } {
# Read bonds and calculate the displacement difference (or disregistry) vector ddrij
MD++ setnolog
initmd " "
# Will be used to tag a bond if its |ddrij| > rc
set rc [expr sqrt(2)/2*$a_Si]
MD++ RLIST = 4.1490
# Read bond data
MD++ incnfile = bonds.dat read_bonds
# The deformed configuration
MD++ incnfile = ../../structures/Examples/deformed-testdisreg.cn readcn eval
# The reference configuration
MD++ incnfile = ../../structures/Examples/ref-testdisreg.cn
MD++ input = $rc calddrij
# Write disregistry data:
# [ddrij.disreg.x ddrij.disreg.y ddrij.disreg.z ddrij.norm ddrij.tag]
MD++ finalcnfile = ddrij.dat write_ddrij
MD++ quit
} elseif { $status == 3 } {
# Read/Plot ddrij
MD++ setnolog
initmd " "
MD++ RLIST = 4.1490
MD++ incnfile = ../../structures/Examples/ref-testdisreg.cn readcn eval
MD++ incnfile = bonds.dat read_bonds
MD++ incnfile = ddrij.dat read_ddrij
fix_atoms_on_tilted_plane 0.0 0.0 -0.095 8
MD++ reversefixedatoms removefixedatoms
setup_window
MD++ slip_normal = \[ -1.0 0.0 1.414214 \]
MD++ plot_limits = \[ 1 -1 1 -1 1 -0.150 -0.031 \]
MD++ openwin alloccolors rotate saverot eval plot_ddrij
MD++ sleep quit
} else {
puts "unknown status = $status"
MD++ quit
}